ACHAIKI IATRIKI | 2022; 41(1):13–17
Editorial
Evanthia Tourkochristou1,2, Athanasia Mouzaki1,3
1Division of Hematology, Department of Internal Medicine, Medical School, University of Patras, Patras, Greece
2Division of Gastroenterology, Department of Internal Medicine, Medical School, University of Patras, Patras, Greece
3Laboratory of Molecular Diagnosis of Infectious Agents, Medical School, University of Patras, Patras, Greece
Received: 14 Jan 2022; Accepted: 01 Feb 2022
Key words: SARS-CoV-2, Omicron variant, COVID-19
Introduction
The continued evolution of SARS-CoV-2, including the rapid accumulation of viral mutations to such an extent that new viral variants with different characteristics are emerging, has led to great concern about the ability of these variants to evade the immune response triggered by natural infections and/or vaccination. A large recent wave of infections has been caused by a new variant classified as B.1.1.529/Omicron. It was first reported by the Network for Genomics Surveillance in South Africa to WHO and classified as a SARS-CoV-2 variant of concern (VOC) on November 26, 2021 [1]. The Omicron variant has four sublineages B.1.1.529 (BA.1), B.1.1.529.1 (BA.1.1), B.1.1.529.2 (BA.2) and B.1.1.529.3 (BA.3), which differ in the number of mutations in the spike protein, with BA.1 having the highest number of mutations and currently being the dominant lineage [2] (Table 1).
The evolutionary history of Omicron is currently unknown because it is very different from the other SARS-CoV-2 variants and may have previously split from other variants, probably Alpha and Delta. Phylogenetic studies suggest that the emergence of Omicron may be related to immunocompromised individuals (e.g., HIV patients co-infected with SARS-CoV-2) harboring Omicron over a period of time, or it may be a host-spawning effect involving an evolutionary pathway in non-human species [3, 4]. The rapid international spread of the Omicron variant and its higher number of mutations compared to other variants, as well as the fact that some Omicron mutations are associated with escape from vaccine-induced immunity, pose a new challenge in the control and prevention of the COVID-19 pandemic [5].
Important questions have been raised about the impact of Omicron on transmissibility, disease severity, the effectiveness of existing COVID-19 vaccines in preventing severe disease, humoral response, and the role of T-cell immunity in vaccinated individuals.
The genetic profile of the Omicron variant and its impact on SARS-CoV-2 characteristics
The Omicron variant has the highest number of mutation sites compared with other SARS-CoV-2 variants, half of which were identified in the spike glycoprotein (S). In total, more than 60 substitutions, deletions, and insertions were reported in the Omicron variant. In the open reading frame genes (ORF) encoding the nonstructural proteins of SARS-CoV-2, the Omicron variant has 9 amino acid substitutions and 3 amino acid deletions. It has been speculated that the deletion of 3 amino acids in ORF1a at sites L3674, S3675, and G3676 may prevent the ability of infected cells to degrade viral components, which would contribute to evasion of the innate immune system [6].
Substitutions and deletions were also dicovered in the structural proteins of SARS-CoV-2, including the envelope, membrane, and nucleocapsid (N) proteins [7]. Two mutations in the N protein, R203K and G204R, have been associated with increased subgenomic RNA expression and viral load [8, 9].
At least 30 amino acid substitutions, 3 small deletions and 1 short insertion of 3 amino acids were identified in the S protein, of which 15 mutations are located in the S1 receptor binding domain (RBD) of the S protein. Mutation D614G in the S protein, common to all SARS-CoV-2 variants, has been associated with higher viral load in the upper respiratory tract [10, 11]. Increased binding between spike and angiotensin-converting enzyme 2 (ACE2) may be due to the N501Y mutation in the S protein, which is present in the Omicron, Alpha, Beta, and Gamma variants and, in combination with the H69/V70 deletion, leads to higher transmissibility [12, 13].
Computer modeling has confirmed that the Omicron variant mutations cause tighter binding at the ACE2-RBD interface, strengthening hydrogen bonds and increasing the accessible surface area [14]. The Omicron variant mutations have also been associated with an increased electrostatic potential of the S protein at the RBD interface with ACE2 compared to the Delta variant, which could lead to a stronger S-ACE2 interaction affinity since ACE2 has a negative electrostatic surface potential. Considering that the entry of SARS-Cov-2 into host cells depends on the binding of RBD to ACE2, the tighter binding of RBD to ACE2 and the stronger RBD-ACE2 interaction affinity caused by the mutations of the Omicron variant might increase the infectivity of SARS-Cov-2 in the upper airway epithelium. The interaction of the S protein with other macromolecules, such as antibodies, could also be affected by the altered electrostatic surface potential of Omicron RBD [15]. By mutating N679K and P681H near the furin cleavage site, Omicron could promote enhanced fusion and infectivity. The amino acid substitutions near the furin cleavage site could insert amino acids that facilitate cleavage of the spike into S1 and S2 and promote fusion between the virus and the host cell membrane [7]. Examination of the changes in the RBD mutations of Omicron in the binding free energy (BFE) of the S-ACE2 protein complex revealed a 10-fold increase in the contagion capacity of Omicron compared to the original SARS-CoV-2 strain and a 2-fold increase in the contagion capacity of Omicron compared to the Delta variant, mainly due to the RBD mutations N440K, T478K, and N501Y [16] (Figure 1).
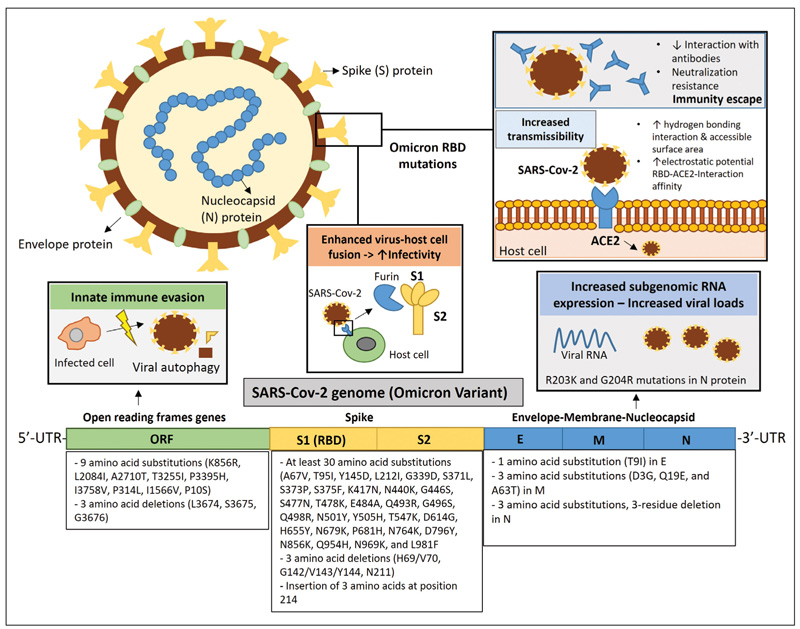
Figure 1. The genetic profile of the Omicron variant and its implications for the viral properties of SARS-CoV-2. The Omicron variant has more than 60 substitutions, deletions, and insertions located in open reading frame genes (ORF), spike glucoprotein (S), envelope protein (E), membrane protein (M), and nucleocapsid (N) protein. Deletion of 3 amino acids in ORF1a at sites L3674, S3675, and G3676 is thought to prevent the ability of infected cells to degrade viral components, contributing to evasion of the innate immune. Two mutations in N protein, R203K and G204R, have been associated with increased subgenomic RNA expression and viral load. Enhanced binding between spike and angiotensin-converting enzyme 2 (ACE2), leading to higher transmissibility was attributed to Omicron mutations in the S1 receptor-binding domain (RBD). According to computer modeling, the Omicron variant mutations could increase the hydrogen bonding interaction and increase the buried surface area accessible to the solvent. Omicron variant mutations were also associated with an increased electrostatic potential of the S protein at the RBD interface with ACE2, which could lead to a stronger interaction affinity of S-ACE2. The interaction of the S protein with antibodies could also be affected by the altered electrostatic potential of the Omicron RBD surface. Structural and clinical studies have shown that Omicron exhibits strong resistance to neutralization by antibodies, indicating the risk of immune escape. Increased infectivity may be favored by Omicron because it has mutations near the furin cleavage site that facilitate cleavage of the spike into S1 and S2, promoting virus-host cell membrane fusion.
The ability of the Omicron variant to escape immunity
Omicron has accumulated a higher number of mutations in RBD compared to Delta [7]. Considering that RBD is the main target for neutralizing antibodies, great concern has been expressed about the ability of Omicron to escape immune recognition and whether existing antibody treatments and vaccines could still be effective.
A clue to the possible resistance of Omicron to neutralization by antibodies was provided by structural analysis studies, which indicated that certain mutations (G446S, Q493R, and G496S) at the S-RBD could create steric interference for antibody binding to the RBD, while other mutations (E484A and Y505H) could abolish interaction affinity with antibodies [17]. The RBD mutations K417N, E484A, and Y505H were held responsible for the high potential of Omicron to interfere with the binding of about 185 antibodies with the S protein [16].
As for monoclonal antibody (mAb) treatment, Omicron’s RBD mutations are thought to interfere with the efficacy of Eli Lilly’s mAb cocktail (against K417N, E484A, Q493R), Celltrion’s regdanvimab antibody (against E484A, Q493R, and Q498R), and the Rockefeller University mAbs (against E484A), whereas the impairment of the efficacy of Regeneron’s mAb cocktail is thought to be low [16].
Some studies have also examined the resistance of Omicron to neutralization by antibodies. In 3 longitudinal cohorts, 169 plasma samples were collected from convalescent individuals who had received or not received 2 doses of Pfizer/BNT or Moderna mRNA vaccine, from uninfected individuals who had received 3 doses of Pfizer/BNT mRNA vaccine, and from uninfected individuals who had received J&J Ad26 vaccine at approximately 1, 5-6, and 12 months after initial vaccination or infection. Neutralizing antibody titers were measured in plasma samples with pseudotyped virus containing the original Wuhan-hu-1 strain or the Omicron variant or a laboratory-developed neutralization-resistant SARS-CoV-2 spike (PMS20). In plasma from convalescent, 2-dose vaccinated and nonvaccinated individuals, neutralization activity was reduced 30- to 60-fold against PMS20 and Omicron compared to the original strain, and the reduced neutralization was even more pronounced in plasma from recipients of 2 mRNA vaccine doses (30- to 180- fold less effective compared with Wuhan-hu-1). In contrast, administration of additional mRNA vaccine doses to infected individuals or those vaccinated with 2 mRNA doses resulted in a 38- to 154-fold and 35- to 214-fold increase in neutralizing activity against Omicron and PMS20, respectively [18].
Decreased neutralizing ability of other recent vaccines against Omicron, including the inactivated virus vaccine BBIBP-CorV and the recombinant dimeric RBD vaccine ZF2001, has also been reported [19]: Neutralization tests were performed using plasma from 37 participants divided into 4 groups: (1) 7 participants after 3-4 months of past SARS-CoV-2 breakthrough infection caused by the Delta variant who had been immunized with 2 doses of the inactivated Sinovac-CoronaVac vaccine before infection, (2) 10 participants who had been immunized with 2 doses of BBIBP-CorV, (3) 10 participants who received a third homologous booster dose of BBIBP-CorV, and (4) 10 participants who received a third heterologous booster dose of BBIBP-CorV/ ZF2001. A plasma pseudovirus neutralization test (pVNT) was performed that included pseudotyped viruses with prototype virus, Beta, Delta, and Omicron variants. Fourteen days after administration of 2 doses of inactivated vaccines, the pVNT titer was lowest for the Omicron variant in 80% of the samples. However, after booster vaccination, positive neutralization sensitivity for the Omicron variant was observed in 100% of the samples.
Although Omicron may have the ability to evade humoral immunity, there are encouraging data on the effect of Omicron on T-cell immunity. Redd et al [20] investigated whether epitopes of the Omicron variant recognized by CD8+ T cells were significantly mutated in recovered COVID-19 patients. The identification of only 1 mutation (T95I) in the spike protein that overlapped with a CD8+ T-cell epitope (GVYFASTEK) associated with two HLA alleles (HLA*A03:01 and HLA*A11:01), and the demonstration of T-cell reactivity in individuals carrying this modified epitope suggests that previously induced anti-SARS-CoV-2 CD8+ T-cell responses may still be active against Omicron and that extensive T-cell escape mutations have not yet developed in SARS-CoV-2.
In addition, a recent analysis of clinical and epidemiologic data from 69,279 SARS-CoV-2-positive patients, 52,297 with S gene drop out (SGTF, presumably the Omicron variant) and 16,982 without SGTF (presumably the Delta variant), found that SARS-CoV-2 infections with the Omicron variant were associated with a substantially lower risk of severe clinical endpoints and shorter length of hospital stay. (Lewnard et al., Pre-print. MedRxiv. 2022. https://doi.org/10.1101/2022.01.11.22269045). However, we do not yet know how infection with all Omicron sublineages affects disease progression.
Overall, Omicron may be better able to evade immune recognition compared with the original SARS-CoV-2 strain and other variants. Although booster vaccination may show promise in restoring the observed reduced neutralizing capacity against Omicron, evaluation of the impact of the third (or fourth) booster vaccination on vaccine efficacy is ongoing because very few studies have been published to date that have examined the neutralizing capacity of various homologous and heterologous booster vaccines against Omicron. It appears that Omicron will not be the last variant, so the development of monovalent vaccines may no longer be the solution. Understanding the specific virology and biology of each new variant is essential to monitor the dynamics of genetic changes and translate this knowledge into more effective prevention strategies for the COVID-19 pandemic.
Acknowledgments
ET is a recipient of KARATHEODORIS grant #80672 from the University of Patras.
Conflict of interest disclosure
None to declare
Declaration of funding sources
None to declare
Authors’ contributions
ET and AM designed and coordinated the study, performed the literature search and analysis, and wrote the manuscript. Both authors approved the submitted version of the manuscript.
References
1. WHO. Enhancing response to Omicron SARS-CoV-2 variant: Technical brief and priority actions for Member States. https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states. 2022
2. Desingu PA, Nagarajan K, Dhama K. Emergence of Omicron third lineage BA.3 and its importance: J Med Virol. 2022; (in press). https://doi.org/10.1002/jmv.27601
3. Kupferschmidt K. Where did ‘weird’ Omicron come from? Science. 2021; 374(6572):1179.
4. Kumar S, Thambiraja TS, Karuppanan K, Subramaniam G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J Med Virol. 2021; 15(10):27526.
5. Cong Z, Evans JP, Qu P, Faraone J, Zheng Y-M, Carlin C, et al. Neutralization and Stability of SARS-CoV-2 Omicron Variant. Pre-print. bioRxiv. 2021. https://doi.org/10.1101/2021.12.16.472934
6. Benvenuto D, Angeletti S, Giovanetti M, Bianchi M, Pascarella S, Cauda R, et al. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J Infect. 2020; 81(1):e24-7.
7. He X, Hong W, Pan X, Lu G, Wei X. SARS-CoV-2 Omicron variant: Characteristics and prevention. MedComm. 2021; 2(4):838-45.
8. Leary S, Gaudieri S, Parker MD, Chopra A, James I, Pakala S, et al. Generation of a Novel SARS-CoV-2 Sub-genomic RNA Due to the R203K/G204R Variant in Nucleocapsid: Homologous Recombination has Potential to Change SARS-CoV-2 at Both Protein and RNA Level. Pathog Immun. 2021; 6(2):27-49.
9. Mourier T, Shuaib M, Hala S, Mfarrej S, Alofi F, Naeem R, et al. Saudi Arabian SARS-CoV-2 genomes implicate a mutant Nucleocapsid protein in modulating host interactions and increased viral load in COVID-19 patients. Pre-print 2021. http://hdl.handle.net/10754/669344
10. Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021; 592(7852):116-121.
11. Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O’Toole Á, et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021; 184(1):64-75.
12. Yang T-J, Yu P-Y, Chang Y-C, Liang K-H, Tso H-C, Ho M-R, et al. Impacts on the structure-function relationship of SARS-CoV-2 spike by B. 1.1.7 mutations. Pre-print. bioRxiv. 2021. https://doi.org/10.1101/2021.05.11.443686
13. Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021; 26(1):pii=2002106.
14. Lupala CS, Ye Y, Chen H, Su XD, Liu H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem Biophys Res Commun. 2021; 590:34-41.
15. Pascarella S, Ciccozzi M, Bianchi M, Benvenuto D, Cauda R, Cassone A. The electrostatic potential of the Omicron variant spike is higher than in Delta and Delta-plus variants: A hint to higher transmissibility? J Med Virol. 2021; (in press). https://doi.org/10.1002/jmv.27528.
16. Chen J, Wang R, Gilby NB, Wei G-W. Omicron (B.1.1.529): Infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. 2022; 62:412-422.
17. Kannan SR, Spratt AN, Sharma K, Chand HS, Byrareddy SN, Singh K. Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J Autoimmun. 2022; 126:102779.
18. Schmidt F, Muecksch F, Weisblum Y, Da Silva J, Bednarski E, Cho A, et al. Plasma neutralization properties of the SARS-CoV-2 Omicron variant. Pre-print. MedRxiv. 2021. https://doi.org/10.1101/2021.12.12.21267646
19. Ai J, Zhang H, Zhang Y, Lin K, Wu J, Wan Y, et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg Microbes Infect. 2021; 22:1-24.
20. Redd AD, Nardin A, Kared H, Bloch EM, Abel B, Pekosz A, et al. Minimal cross-over between mutations associated with Omicron variant of SARS-CoV-2 and CD8+ T cell epitopes identified in COVID-19 convalescent individuals: Pre-print. bioRxiv. 2021. https://doi.org/10.1101/2021.12.06.471446